-
REAGENT SERVICES
Hot!
-
Most Popular Services
-
Molecular Biology
-
Recombinant Antibody/Protein
-
Reagent Antibody
-
CRISPR Gene Editing
-
DNA Mutant Library
-
IVT RNA and LNP Formulations
-
Oligo Synthesis
-
Peptides
-
Cell Engineering
-
- CRISPR/Cas9 sgRNA
- CRISPR/Cas12a crRNA
- Prime Editing Guide RNA
- Base Editing Guide RNA
- HDR Templates
- gRNA + HDR Template Design Tools
- cGMP Guide RNA
- cGMP HDR Templates
- CRISPR/Cas Proteins
- CAR-T Knock-in Optimization Kit
- CRISPR Plasmids
- CRISPR gRNA Plasmid Libraries
- CRISPR Cell Lines
- Microbial Genome Editing
-
-
PRODUCTS
-
Most Popular Products
-
Antibodies
-
ELISA Kits
-
Protein Electrophoresis and Blotting
-
Protein and Antibody Purification
-
Recombinant Proteins
-
Molecular Biology
-
Stable Cell Lines
-
Cell Isolation and Activation
-
 IVD Raw Materials
IVD Raw Materials
-
 Therapy Applications
Therapy Applications
-
Online Shop
-
Resources
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
![Anti-Camelid VHH]() MonoRab™ Anti-VHH Antibodies
MonoRab™ Anti-VHH Antibodies
-
![Precast Gels]() SurePAGE™ Precast Gels
SurePAGE™ Precast Gels
-
![Quatro ProAb Automated Protein and Antibody Purification System]() AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
AmMag™ Quatro ProAb Automated Protein and Antibody Purification System
-
![Target Proteins]() Target Proteins
Target Proteins
-
![AmMag™ Quatro Automated Plasmid Purification]() AmMag™ Quatro automated plasmid purification
AmMag™ Quatro automated plasmid purification
-
 IVD Raw Materials
IVD Raw Materials
-
![Quick
Order]() Quick Order
Quick Order
-
![Quick
Order]() Quick Order
Quick Order
- APPLICATIONS
- RESOURCES
- ABOUT US
- SIGN IN My Account SIGN OUT
- REGISTER

![Protein Analytical Service Protein Analytical Service]()
Protein Analytical Service
Empowering High-Quality Antibody Production with Advanced Protein Analysis Technology
Home » Protein Expression » Protein Analytical Service
Overview
GenScript’s Comprehensive Protein Analysis Platform is an advanced, integrated system designed for in-depth study and evaluation of protein characteristics. It offers a wide range of services, including qualitative & quantitative analysis, protein modification analysis, impurity analysis, molecular interaction analysis, and more customized analysis. Together, these capabilities make the platform an essential tool in protein research, drug development, and biopharmaceutical production.

Qualitative & Quantitative Analysis
- Concentration
- Purity (SDS-PAGE, CE-SDS, icIEF, SEC-HPLC, RP-HPLC, HIC-HPLC, etc.)
- Intact mass /Native mass
- (in gel) Peptide mapping
- N/C terminal sequencing

Protein Modification Analysis
- PTM analysis
- Disulfide bond mapping
- N-glycan and O-glycan profiling
- Sialic acid analysis
Impurity
Analysis- Endotoxin quantification
- HCP identification /quantification
- Residual DNA quantification
- Residual Protein A quantification

Molecular Interaction
Analysis- Antibody-antigen affinity analysis >>
- Protein-protein/DNA /small molecule interactions
- Fc effector function
- Enzyme activity assay

Customized
Analysis- Monoclonal antibody self-interactions
- Monoclonal antibody polyspecificity
- Thermal stability analysis
- Mutation site analysis
- Free-thaw stability analysis
- Antibody de novo sequencing
- More customized services available
Advanced instruments for protein analysis

Waters BioAccord LC-MS

Thermo QE Orbitrap LC-MS/MS

Waters Xevo G3 QTOF

Biacore 8K

Octet RED
qPCR

Plate reader

TR-FRET

LabChip CE-SDS

Maurice iCIEF

Agilent GC

Cedex analyzer

Waters UPLC
Optimize Your Research with Precision-Fit Assays
Identify your research stage and discover tailored services to accelerate your progress!
 Antibody Drug DiscoveryGene to HitsHits to LeadsCandidates SelectionDiscovery & Target
Antibody Drug DiscoveryGene to HitsHits to LeadsCandidates SelectionDiscovery & Target
IdentificationHits ScreeningLead Indentifaction
& ValidationLead OptimizationIn vivo & vitro AssaysGLP Tox0.05mg500mg1g200gTurboCHO™ HTTurboCHO™ ExpressTurboCHO™ HP/TurboCHO™ StableQualitative & Quantitative Analysis
Molecular Interaction Analysis- Purity by CE-SDS or HPLC
- Affinity screening/ranking >>
- HT antibody stability screening
- More customized services available
Qualitative & Quantitative
Analysis- Purity by HPLC
- Intact mass
- Peptide mapping
- N/C terminal sequencing
- More customized
services available
Protein Modification & Molecular
Interaction Analysis- Identification by mass
- PTM analysis
- Disulfide bond mapping
- N-/O- glycan profiling
- Sialic acid analysis
- Fc binding affinity >>
- More customized services available
 In Vitro Diagnostic DevelopmentTarget ScreeningAssay DevelopmentAssay Validation and OptimizationDiagnostic
In Vitro Diagnostic DevelopmentTarget ScreeningAssay DevelopmentAssay Validation and OptimizationDiagnostic
CommercializationBiomarker
DiscoveryReplication
Confirm markers and reproducibility of assaysAnalytical Development
Translation onto a clinical diagnostic platformPhase I
Sensitivity & SpecificityPhase II
PPV and NPVPhase III
Clinical benefit
& Cost effectivenessRegistration0.05mg500mg1g10gTurboCHO™ HTTurboCHO™ ExpressTurboCHO™ HP/TurboCHO™ StableTurboCHO™ StableQualitative & Quantitative Analysis
Molecular Interaction- Purity by CE-SDS or HPLC
- Affinity screening/ranking >>
- More customized services available
Protein Modification & Molecular Interaction Analysis
- Identification by mass
- PTM analysis
- Glycan analysis
- Binding affinity assay >>
- More customized services available
Interested in learning more about protein assay method?
Discover their applications, benefits, and explore case studies below.
-
Qualitative & Quantitative Analysis
-
Protein Modification Analysis
-
Molecular Interaction Analysis
-
Impurity Analysis
Qualitative & Quantitative Analysis Applications Assay list Benefits Purity Analysis SDS-PAGE - High acceptance
CE-SDS - High accuracy
- Automated
- High resolution
HPLC - Broad applicability
UPLC - Ultra-high pressure resistance
- Minimal delay volume
- Minimal sample residue
Routine molecular weight detection Non-deglycosylated detection:
- Intact mass
- Reduced mass
- Suitable for characterization of glycosylation modifications and protein assembly
N-glycan removal detection:
- De-glycosylated mass
- Reduced and de-glycosylated mass
- High intensity mass spectrometry
- More accurate determination of protein chain
- Suitable for bispecific antibody identification
Characterization of heavily glycosylated proteins Denatured N-glycan removal detection:
- De-glycosylated mass(denatured)
- Reduced and de-glycosylated mass(denatured)
- Suitable for the identification of complete molecular weight measurement of proteins with complex modifications.
Denatured N-glycan and O-glycan removal detection:
- De-O-glycosylated mass
- Reduced and de-O-glycosylated mass
- Simultaneous removal of N-glycans and O-glycans
- Elimination of their interference with protein detection
Characterization of complex formation under the native state - Native mass
- Mild detection conditions and native state preservation
- Suitable for analysis of protein complexes linked by non-covalent bonds
- Suitable for analyzing the degree of small protein aggregation
Characterization of the complete protein sequence - Sequence coverage
- Minimal impact of protein purity
- 100% sequence coverage
- in-gel sequence coverage
- Qualitative analysis of the band of interest
Characterization of protein N-terminal and C-terminal sequences - N/C-terminal sequence
- Confirm full protein expression
- Detect breaks in protein expression
- Identify N/C-terminal modifications
Case1 Native mass - BsAb – Identification of the non-covalently linked chain
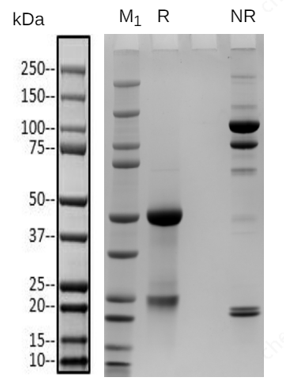
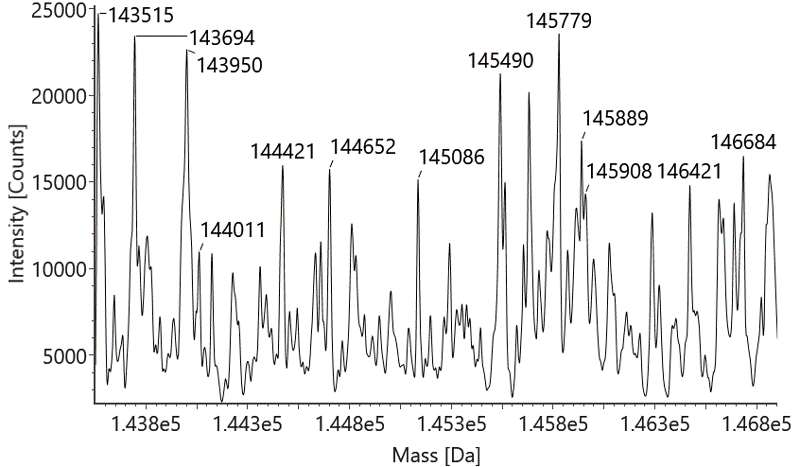
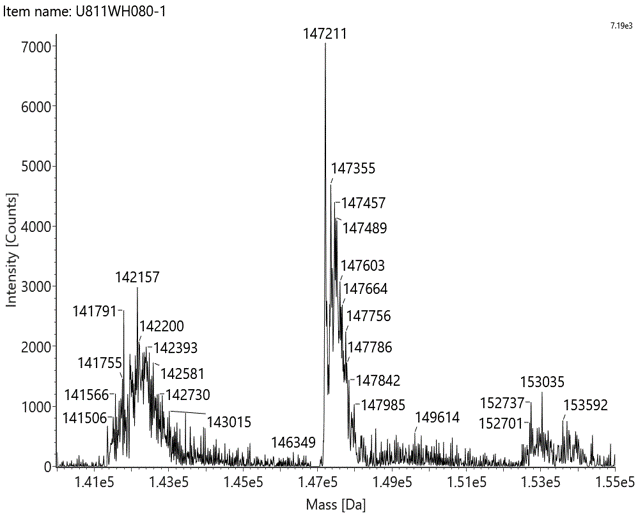
Native mass spectrometry is often used to identify the components of bispecific antibodies. The molecular weights of the marker bands in Fig 1A are 150 kDa, 120 kDa, 100 kDa, 74 kDa, and 23 kDa. Under denaturing conditions, the 150 kDa light chain was not identified by conventional mass spectrometry. This indicates that non-covalent interactions between proteins were disrupted under these conditions (Fig 1B). In contrast, native mass spectrometry reflected the original state of the proteins and successfully identified the light chain (Fig 1C).
Figure 1 SDS-PAGE and Mass analysis of BsAb1
A
B
C
Case 2 In-gel sequence coverage - Biotinylated protein – Identification for low concentration protein
Detection of biotinylation efficiency for protein-1 was requested by the customer. However, its concentration is far below the requirement and it is difficult to concentrate. Nevertheless, the SDS-PAGE bands were relatively clear (Fig 1). By excising the target band from the gel and performing coverage analysis, the biotinylation efficiency was successfully identified (Fig 2, Tab 1).
Figure 1 SDS-PAGE of protein-1
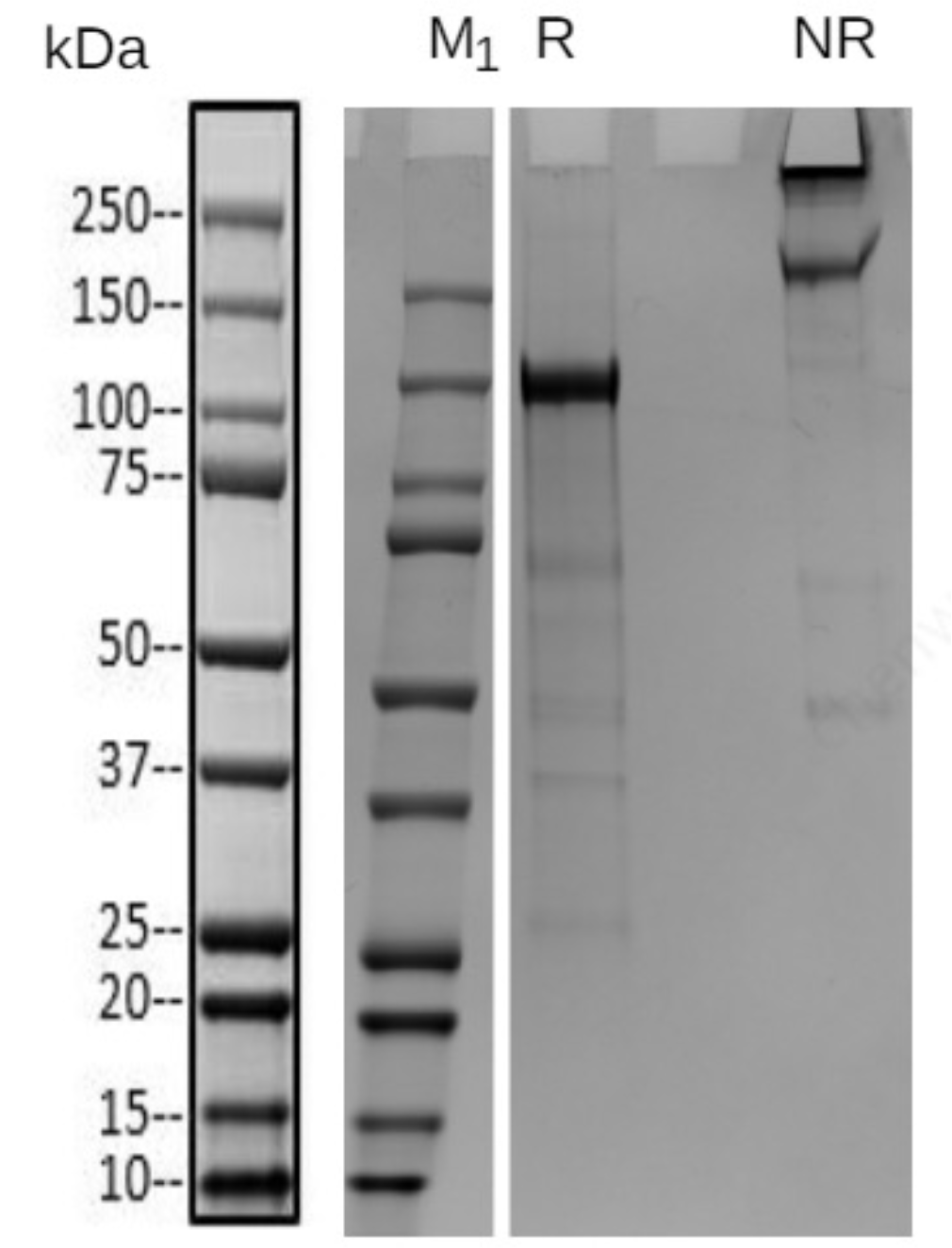
Figure 2 Fragment coverage map of protein-1
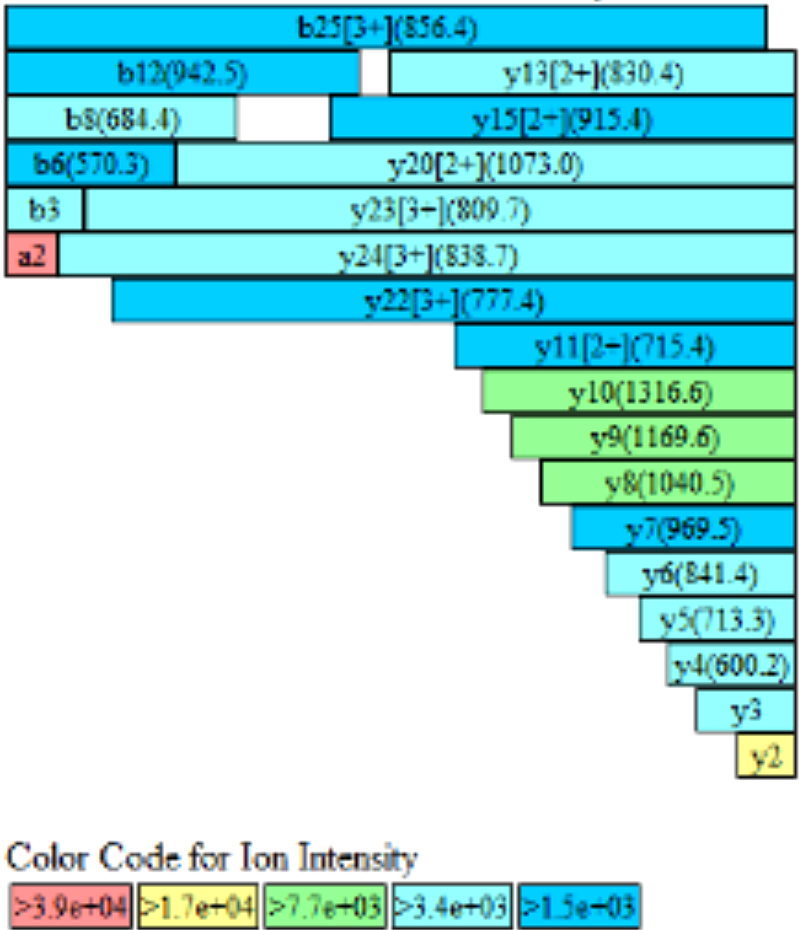
Table 1 Peptide coverage analysis of protein-1
Site Mod M/Z Charge St. Mono Mass Exp. Mono Mass Theo. △ ppm Biotinylation ratio % K1296 Biotinylation 980.803 3 2938. 385 2938.392 -2.36 99.3 Protein Modification Analysis Assay List Benefits Glycosylation mirror comparison - Suitable for the analysis and determination of glycosylation sites and their respective proportions in proteins.
Biotinylation ratio
(peptide mapping)- Targeted enzymatic cleavage for Avi tag analysis
- High accuracy
Disulfide bond analysis - Suitable for determination of sites and proportions of disulfide bond formation between cysteine residues in proteins
N-glycosylation site - Suitable for determination of sites and proportions of N-glycosylation
PTM-phosphorylation - Suitable for determination of sites and proportions of protein modifications
PTM-deamidaton PTM-oxidation Case1 Glycosylation mirror comparison – Glycoprotein – Production process optimization and selection
Ab-1 and Ab-2 are the same protein produced by different hosts with different glycosylation patterns. Glycosylation mapping analysis identifies the glycosylation sites and proportions of Ab-1 and Ab-2, facilitating process optimization and selection.
Figure 1 Reduced Mass analysis of Ab-1 & Ab-2
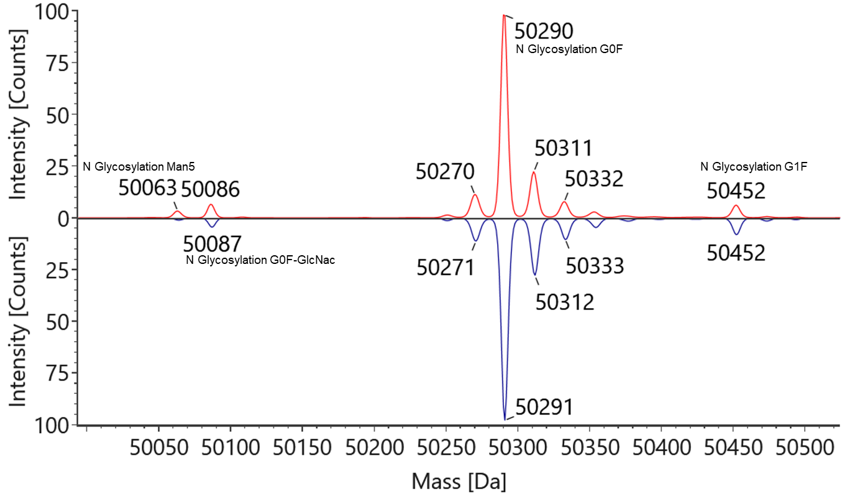
Table 1 Glycosylation analysis of Ab-1 & Ab-2
NO. Mass (Da) Glycan Name Ratio% Ab-1 Ab-2 1 50063 N Glycosylation Man5 3.3% 1.4% 2 50086 N Glycosylation G0F-GlcNac 5.9% 4.2% 3 50290 N Glycosylation G0F 84.5% 85.9% 4 50452 N Glycosylation G1F 6.3% 8.6% Note:Mass value is from Ab-1.
Molecular Interaction Analysis Applications Assay List Benifits Recommendation - Antigen-antibody affinity analysis
- Protein-protein interaction
- Protein-DNA interaction
- Protein-small molecule interaction
Bio-Layer Interferometry >> - Target-free, real-time detection, high throughput
- Relatively low cost
- High sample compatibility
- Sample recovery available
- Suitable for the detection of supernatants
- BLI and SPR perform comparably when used to detect samples with moderate affinity(KD µM~nM).
Surface Plasmon Resonance >> - Target-free, real-time detection, high throughput
- High sensitivity, broader affinity detection range
- Capable of detecting small molecules (100 Da)
- Suitable for detecting low-affinity(KD ~µM) and higher affinity (KD ~0.1 nM) samples
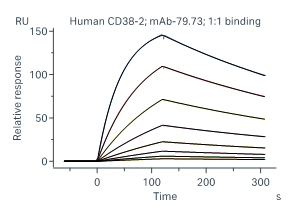
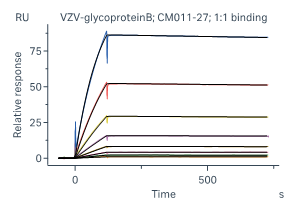
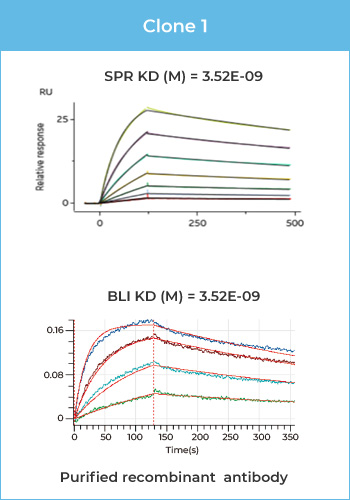
Case 1 SPR – IgG Affinity –SPR has high sensitivity and broad affinity detection range (KD = 10-3-10-15)
As shown in Fig 1 and Tab 1, SPR has higher sensitivity and a wider affinity detection range than BLI. SPR is suitable for both low-affinity (KD ~µM) and high-affinity (KD ~0.1 nM) samples, which may not be successfully detected by BLI.
Figure 1 Affinity binding curves of human IgG-1, 2, 3
Human IgG-1
Human IgG-2
Human IgG-3
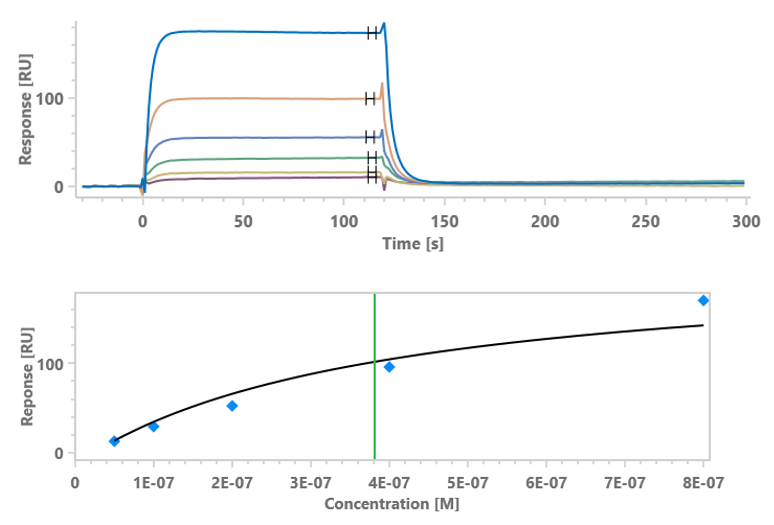
Table 1 Affinity binding detailed analysis report of human IgG-1, 2, 3
Ab Chip Affinity Test item Chi² (RU²) Ka (1/Ms) kdis (1/s) KD (M) Human IgG-1 ProA Medium Affinity 4.90e-02 1.39e+05 2.07e-03 1.49e-08 Human IgG-2 ProA High Affinity 3.41e-01 2.75e+04 3.59e-05 1.30e-09 Human IgG-3 ProA Low Kinetics 4.75 - - 3.818E-07 Case2 BLI v.s. SPR – IgG Affinity - BLI and SPR results are comparable for most antibody-antigen interactions
Most antigen-antibody interactions fall into the category of medium affinity samples, for which the detection results (KD) of BLI and SPR are nearly identical.
Rabbit IgG
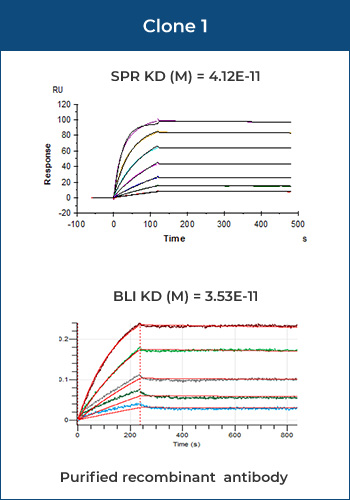
Rabbit IgG
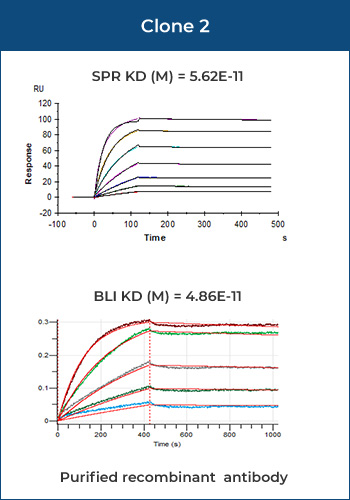
Mouse IgG
Impurity Analysis Applications Test Items Benefits Host cell protein analysis ELISA or LCMS - Provides reference for upstream manufacturing and downstream applications
Residual DNA qPCR based / Residual Protein A ELISA / Case 1 Host cell protein analysis – Impurity protein – SDS-PAGE unknown band Identification
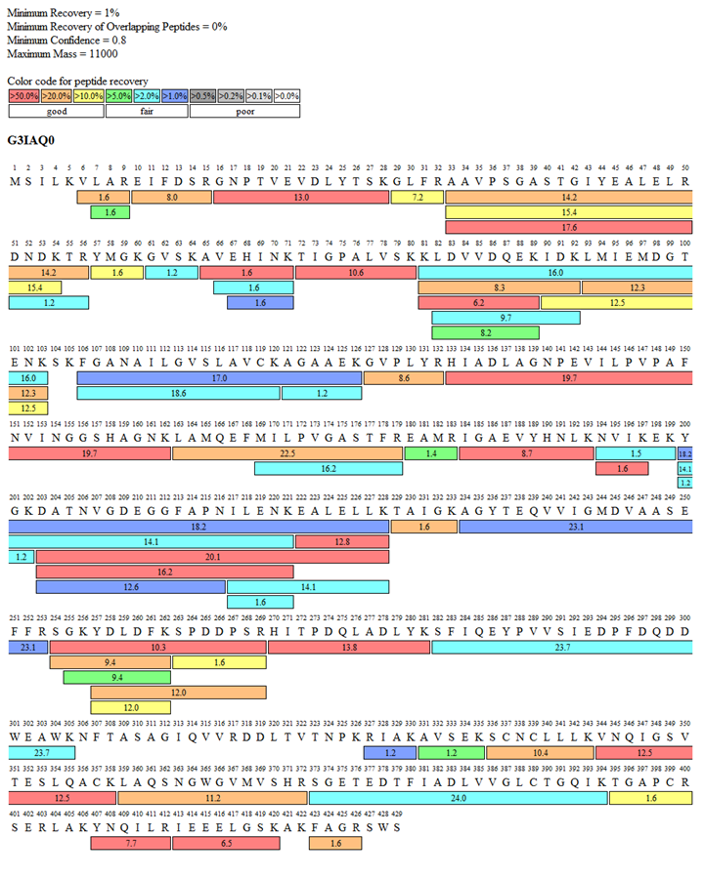
An unknown band was detected in several production batches of the protein (Fig 1). After excluding protein contamination, this band was suspected to be a host cell protein (HCP). HCP analysis preliminarily identified the band as phosphopyruvate hydratase (Tab 1). Further sequence analysis revealed a sequence coverage of 90.9% for this protein, confirming the band as the host cell protein phosphopyruvate hydratase (Fig 2).
Figure 1 SDS-PAGE of expression sample
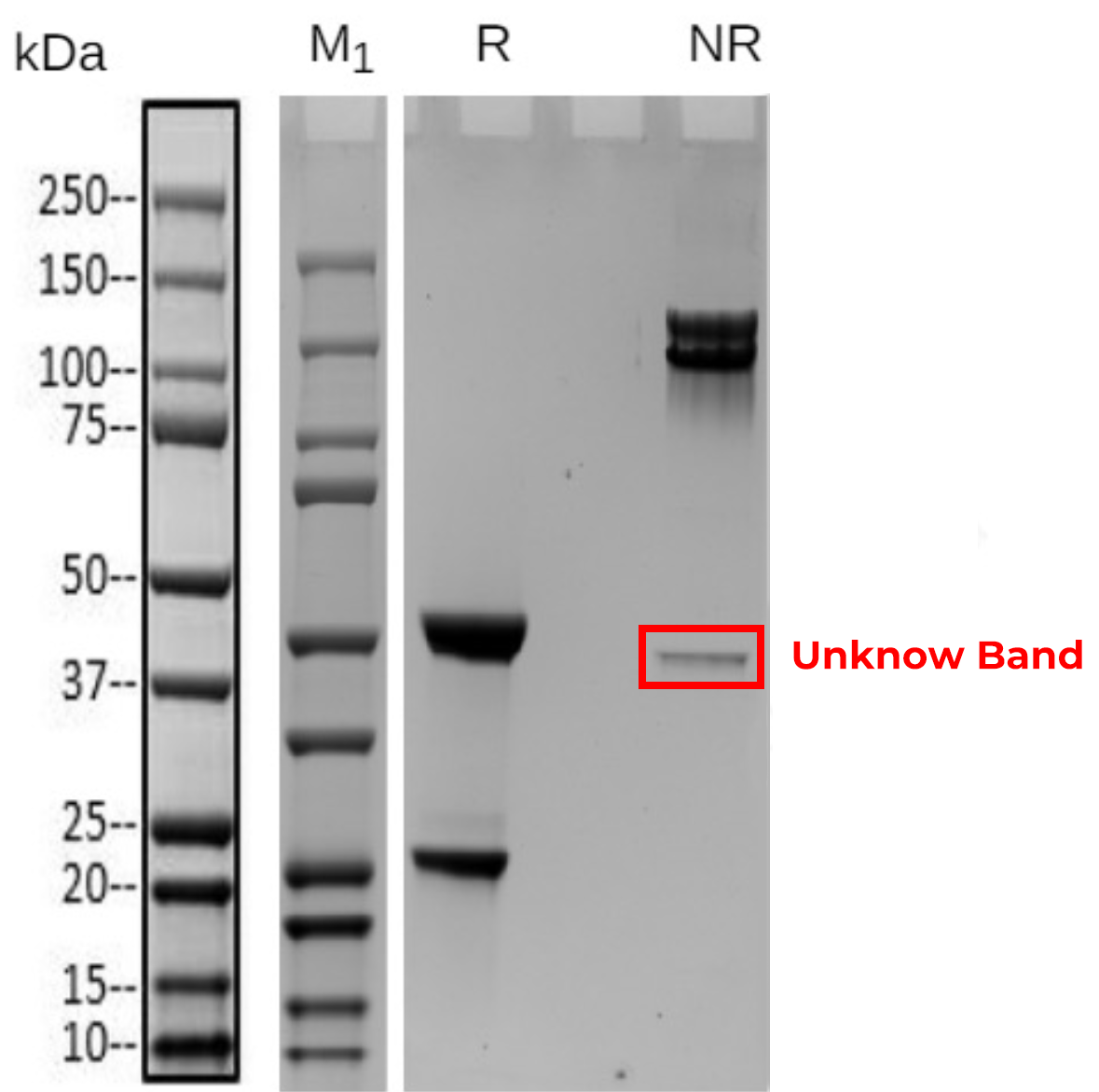
Table 1 Sequence coverage map report of the unkown band
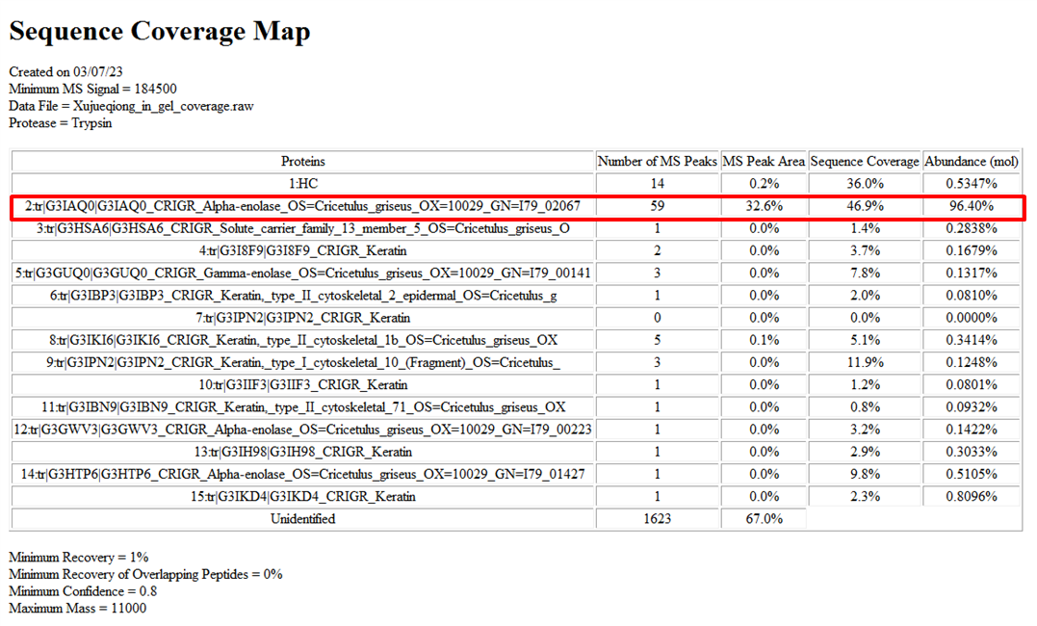
Figure 2 Sequence coverage map of the unkown band
Get in Touch
with GenScript Protein Analytical Service

-


































